| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website http://www.gastrores.org |
Original Article
Volume 11, Number 1, February 2018, pages 46-51
Analysis of Helicobacter pylori Genotypes Amongst Jordanians’ Dental Plaque Samples
Mohammad Abu-Lubada, f, Hamed Alzoubia, Dua’a Jarajreha, Alaa Al Sawalqab, c, Holger Bruggemannd, Eman Albataineha, Amin Aqela, Munir Al-Zeere
aDepartment of Medical Microbiology and Immunology, Mutah University Faculty of Medicine, Karak, Jordan
bRoyal Medical Services, Amman, Jordan
cDepartment of Dentistry, King Hussein Medical Center, Amman, Jordan
dDepartment of Biomedicine, Aarhus University, Aarhus, Denmark
eDepartment of Molecular Biology, Max Planck Institute of Infection Biology, Berlin, Germany
fCorresponding Author: Mohammad Abu-Lubbad, Department of Medical Microbiology and Immunology, Mutah University Faculty of Medicine, P.O. Box 7, Mutah 61710, Karak, Jordan
Manuscript submitted December 14, 2017, accepted January 3, 2018
Short title: H. pylori Genotypes in Dental Plaque Samples
doi: https://doi.org/10.14740/gr947w
| Abstract | ▴Top |
Background: Helicobacter pylori (H. pylori) infection has been associated with gastritis, gastric ulcer, mucosa-associated lymphoid tissue lymphoma and gastric cancer. The prevalence of H. pylori virulence genes have been studied in different populations and from different sources of samples but their prevalence has not been studied in dental plaque in Jordanian people; therefore, the aim of this study was to determine the genotypes of H. pylori isolated from dental plaque samples.
Methods: Dental plaque samples were collected from 60 Jordanian volunteers. The genotypes of H. pylori virulence genes including the cytotoxin-associated gene A (cagA) and the vacuolating toxin (vacA) were determined using polymerase chain reaction (PCR).
Results: The cagA gene was detected in 14 (23.3%) samples, while vacA was detected in all volunteers enrolled in this study (100%). The most prevalent vacA alleles were m2 and s1 in 54 (90%) and 55 (91.7%) of volunteers, respectively. Compared to the other combinations including the most virulent vacA genotype s1/m1 which was detected in 11 (18.2%) of volunteers, the most prevalent vacA allelic combinations were s1/m2 and s2/m2 in 56 (93.3%) and 27 (45%) of volunteers, respectively.
Conclusions: These results indicate a significant carriage of virulent H. pylori strains among Jordanian people in their dental plaques, which increases the possible transmission of these strains among them. In addition, the studying of the genotypic pattern of H. pylori virulence genes in the dental plaque could represent an essential tool for infection prevention and predicting the severity and prognosis of H. pylori gastric infection.
Keywords: Jordanian; Helicobacter pylori; Dental plaque; Virulence genes; The cytotoxin-associated gene; The vacuolating toxin; Alleles; Polymerase chain reaction
| Introduction | ▴Top |
Helicobacter pylori (H. pylori) infection has been found to induce chronic gastritis, gastric ulcer, mucosa associated lymphoid tissue lymphoma, and gastric carcinoma [1]. Gastric carcinoma is categorized as the third most common cause of death worldwide [2]. Bacterial genetic heterogeneity, environmental factors and age of acquisition determine the clinical outcome associated with H. pylori infection [3, 4]. The genetic variation within genes encoding virulence factors plays a major role in the pathogenesis of different H. pylori strains [5]. The virulence genes of H. pylori are located within a DNA segment of 35 - 40 kbp called the pathogenicity island. The cytotoxin-associated gene A (cagA) and the vacuolating toxin (vacA) are the most studied genes among others [6, 7]. They are used as prognostic factors and as epidemiological markers [8]. It has been found that more than 50% of H. pylori isolates carry the cagA gene [9]. Upon infection with H. pylori, the CagA protein is translocated into the cells by a type IV secretion system. CagA as a virulence factor is known to deregulate several vital signaling pathways of the host including the disruption of tight junctions that increases probability of gastric ulceration and cancer development [10].
The vacA gene encodes the vacuolating cytotoxin A which increases the risk of peptic ulceration and gastric cancer [11] through the induction of vacuolation in gastric cells, interfering with mitochondrial functions, and stimulation of apoptosis [12]. Despite the fact that vacA gene has been isolated from all H. pylori strains, allelic variation was found due to polymorphisms in two significant vacA regions. The first region is a region referring to as signal region (s) and the second region located at the middle of the gene called the middle (m) region [11]. Two alleles of the m region, m1 and m2, and two alleles of the signal peptide, s1 and s2 were identified. Within allele s1, subtypes s1a and s1b have been distinguished [1]. Studies have proved that the different combinations of vacA alleles determine the degree of cytotoxicity [13, 14]. The bacterial isolates possessing s1/m1 have higher cytotoxicity, which enhances the possibility of gastric inflammation, atrophy, and gastric carcinoma compared to bacterial isolates with s2/m2 or s2/m1 combinations [2, 13].
The patterns of H. pylori genotypes were identified in different populations from gastric tissues, oral cavities, and other sources worldwide. We previously demonstrated the prevalence of H. pylori in dental plaque samples from Jordanian people [15], the aim of the present study was to determine the patterns of vacA and cagA genotypes of H. pylori from dental plaques among Jordanians.
| Materials and Methods | ▴Top |
Sample collections
A total of 60 Jordanian volunteers were invited to participate in this study as published before [15]. Using sterile curette, dental plaque samples were collected from the dentistry clinic at the medical center of Mutah University. Thirty samples were collected from students and the others were collected from the employees working at Mutah University and other visitors. The ages of the participants ranged between 18 and 52 years old with a mean age of 25. Forty one (68.3%) samples were from man and 19 (31.7%) from woman. All participants were non-smokers, not under antibiotic treatment for the previous 4 weeks of the sample collection, and all showed good general health status including healthy oral cavity, which was important to have similar buccal environment for all collected samples. Samples were collected after signing an informed consent from all participants in this study, which was approved from The Scientific Research Ethical Committee in The Faculty of Medicine at Mutah University under the reference number 20170.
DNA extraction
Genomic DNA from the dental plaque samples was extracted using DNA extraction mini kit (Omega, Bio-Tek, USA) according to the manufacturer’s instructions. The isolated DNA was eluted in 100 µL nuclease free water, quantified on a UV-nano spectrophotometer (Quawell Technology Inc, USA), and stored at -20 °C until tested [15].
H. pylori genotyping for cagA and vacA
Polymerase chain reaction (PCR) was carried out as described previously [1]. In brief, a volume of 50 µL containing 1 µM of each primer, 1 µL of genomic DNA (approximately 200 ng), 1 mM of dNTPs mix (1 µL), 0.5 µL Phusion High-Fidelity DNA Polymerase (Thermo Scientific, USA) were used to detect the presence of H. pylori cagA and vacA s and m regions. PCR amplifications were carried out using Applied Biosystems Thermal PCR Cycler (Thermo Fisher, USA). Table 1 summarized the details of the primers sequences used in this study and Figure 1 showed the PCR results of the amplified genes. After optimization, the following cycle conditions were used for cagA: 35 cycles of 7 s at 98 °C, 30 s at 56 °C, and 30 s at 72 °C, while, the following conditions were used for all vacA primers: 35 cycles of 7 s at 98 °C, 30 s at 53 °C, and 30 s at 72 °C. Negative controls and positive samples were included in each run. Amplified PCR products were then resolved by electrophoresis on 2% agarose gels run and visualized using Thermo Fisher Scientific gel documentation system.
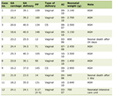 Click to view | Table 1. PCR Primers for Amplification of cagA and vacA Genes |
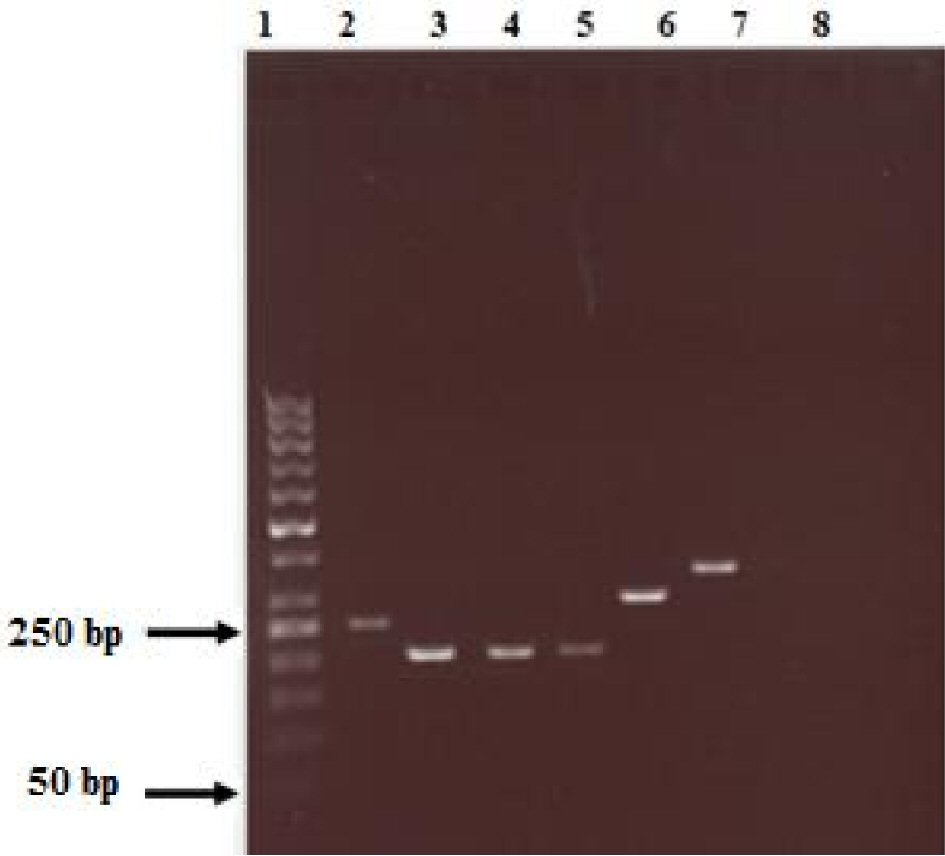 Click for large image | Figure 1. PCR genotyping of vacA and cagA genes from the volunteers’ samples (lanes- 1 = 50 bp molecular weight marker; 2 = cagA+; 3 = S1a; 4 = S1b; 5 = S2; 6 = m1; 7 = m2 and 8 = negative control (without DNA)). |
| Results | ▴Top |
A total of 60 dental plaque samples were analyzed to study the genetic patterns of H. pylori cagA and vacA virulence genes. Previously we had confirmed the presence of H. pylori in all analyzed samples obtained from dental plaques among Jordanian people [15]. In the current study, cagA and vacA genes were analyzed and detected from the same dental plaque samples that were used previously [15]. H. pylori cagA gene was found in 14 samples, which represented 23.3% of the total number of samples. Our data confirmed the presence of the s and m alleles of vacA in all samples (Table 2). The vacA m2 allele which was detected in 54 samples (90%) was more prevalent than vacA m1 allele which was found in nine samples (15%). Only four samples (6.6%) were found to contain m1 and m2 alleles as shown in Table 2, which indicates, probably, the presence of different H. pylori strains. Concerning vacA s region, the s1 allele was found in 91.7% of samples in which 33 samples (55%) were subtype s1a and 32 samples (53.3%) were subtype 1b. Moreover, s2 was found in 30 samples (50%) as shown in Table 2. In this study, the combinations of vacA regions genotypes were identified as follow: s1/m1 (18.2%), s1/m2 (93.3%), s2/m2 (45%), and s2/m1 genotype was found in 10% of our samples (Table 2). These findings clearly suggest the presence of different genotypes of H. pylori in the dental plaques samples among Jordanian individuals.
 Click to view | Table 2. Prevalence of cagA Gene, vacA Alleles, and vacA Allele Combinations |
| Discussion | ▴Top |
H. pylori is considered the most common causative agent of gastric infections reported worldwide, with a prevalence ranging from 25% to 80% in the developed and the developing countries, respectively [16, 17]. H. pylori infection is associated with gastritis, peptic ulcers, and gastric cancer; therefore, H. pylori is classified as a carcinogenic bacterium [1]. It is believed that H. pylori is transmitted among people through the oral cavity. The hypothesis that the mouth is a reservoir for H. pylori and a potential source of gastric infection is supported by several studies proved the presence of H. pylori DNA in the saliva and dental plaque [15, 18-21]. Abu Lubad et al proved that the dental plaque is considered as a potential reservoir for H. pylori and is might be an important source for the gastric infection and reinfection among Jordanian people [15]. To our knowledge, there are no studies conducted on the prevalence of H. pylori vacA and cagA genotypes among the Jordanian population from dental plaque samples. The clinical relevance and geographical distribution of the virulent genotypes of H. pylori is still a matter of debate. Our current study reported the prevalence and genotypes of H. pylori virulence genes cagA and vacA in addition to the allelic variations of the vacA gene from dental plaques from Jordan, in which cagA gene was found in 23.3% of dental plaque samples in this study, while vacA genotype was found in 100% of samples tested.
H. pylori CagA and VacA are the most extensively studied virulence cytotoxins. CagA and VacA toxins are responsible for the progression to more severe disease conditions and the deregulation of many host cell pathways. The genetic distribution of the H. pylori cagA and vacA in different populations has been extensively studied using different sources including gastric biopsies, stool, and saliva samples [22-26]. In this study, we aimed to investigate the genotypic distribution of H. pylori virulence cytotoxins in dental plaque samples among Jordanian people. Surprisingly, the prevalence of cagA gene in dental plaque samples in this study was similar to its distribution in gastric biopsies obtained from Jordanian patients having gastrointestinal diseases [27]. CagA gene was found in 23.3% of the Jordanians’ dental plaque samples which is similar to that in the dental plaque samples among Brazilian and Iranian people (24.1% and 27.3%, respectively) [26, 28]. In British children, cagA gene was detected in six (17%) out of 36 dental plaque PCR positive samples [29]. It is known that cagA is present in 40% to 90% of H. pylori strains in Western countries and in East Asian countries, respectively [24]. In Taiwanese patients, the cagA was positive in 79% and 9% of stomach antrum and body samples, respectively [9]. CagA was also detected in 60%, 61.8%, 87.5%, 61.6%, and 46.7% among Chilean, Saudi, Chinese, Tunisian, and Brazilian patients, respectively [1, 2, 23, 26, 30].
VacA is the other important virulence factor of H. pylori that has a cytotoxic effect and has an ability interfere with important cellular functions [12]. Our study revealed that all dental samples were positive for vacA with variability in the expression of different alleles among different samples. Concerning the s region of vacA, the prevalence of s1a (55%) region was almost similar to that of s1b (53%) and s2 (50%) alleles among Jordanian people. Regarding the m region of vacA, the m2 allele was found in 90% of samples compared to only 15% for m1 allele. One important determinant of the pathogenicity of H. pylori is the presence of certain vacA genotypes. The most pathogenic genotype is s1/m1 which is associated with high levels of vacuolation capacity compared to the moderate effect of s1/m2, while s2/m2 and s2/m1 vacA alleles lack detectable cytotoxin activity [11, 31, 32]. In our study, 93.3% of samples were positive for the s1/m2 genotype, followed by 45%, 10%, and 18.2% for s2/m2, s2/m1, and s1/m1, respectively. These percentages of allelic combinations indicate the presence of mixed serotypes of H pylori in the dental plaque samples among Jordanian people.
On the other hand, a previous report showed that gastritis samples from Jordanian patients showed an equal prevalence of s1/m1 and s2/m2 (46.2%), while 7.7% for s1/m2 and the genotype s2/m1 was not detectable [27]. It has been also demonstrated that the expression of both cagA and vacA s1/m1 genotypes in gastric samples are involved in more severe disease outcome and are associated with an increased risk of developing distal gastric adenocarcinoma [11, 19, 33]. Other reports linked the cagA and vacA s1/m1 genotypes in dental plaque samples with more severe gastric disease [34, 35]. However, in our study we found around 5% of individuals tested in this study are positive for both cagA and vacA s1/m1 genotype and despite this small percentage, those people who are considered a high-risk group for the transmission of a highly virulent strain of H. pylori to other people [11, 19, 33, 35]. Therefore, studying the genotype pattern of vacA gene can be a tool to help in applying vigorous infection prevention measures in endoscopy units and in the management plan by predicting the severity and prognosis of H. pylori infection.
The present study showed that the majority of H. pylori detected in the dental plaque samples carry virulence-associated genes, which might make the oral cavity a potential route for the transmission of the highly virulent H. pylori strains. It has been proved that the H. pylori strains that are associated with stomach reinfection were similar to that present in the oral cavity [36-39]. Because the oral cavity in Jordanian people harbor virulent strains of H. pylori, it seems essential that the diagnosis and therapy described for H. pylori gastric infection individuals accompanied with additional molecular testing for the dental plaque if their gastric H. pylori infection recurs.
In conclusion, our study is the first report with an extensive coverage of the prevalence of the H. pylori virulence-associated genotypes from dental plaque samples among Jordanian people. Our results showed that the majority of H. pylori from dental samples carry multiple virulence genes that might be easily transmitted among Jordanian people causing severe gastric diseases. The presence of highly pathogenic H. pylori in dental plaque samples might be exploited to clarify the reason behind the high prevalence of H. pylori associated gastritis in Jordanian people [27]. Finally, studying the prevalence of virulence-associated H. pylori genotypes would have profound clinical and epidemiological implications in our understanding of the pathogenesis of H. pylori in Jordan.
Acknowledgments
The authors deeply appreciate and thank Prof. Dr. Thomas F Meyer who is a director at Max Planck Institute for Infection Biology/Berlin, for his support to do the genetic analysis in his lab; also, many thanks extend to the German Academic Exchange Service (DAAD) for their financial support. We would like to thank the staff at the dental clinic in Mutah University for helping in samples collection.
Conflict of Interest
The authors declare that they have no competing interest.
| References | ▴Top |
- Marie MA. Relationship between Helicobacter pylori virulence genes and clinical outcomes in Saudi patients. J Korean Med Sci. 2012;27(2):190-193.
doi pubmed - Pinto-Ribeiro I, Ferreira RM, Batalha S, Hlaing T, Wong SI, Carneiro F, Figueiredo C. Helicobacter pylori vacA Genotypes in Chronic Gastritis and Gastric Carcinoma Patients from Macau, China. Toxins (Basel). 2016;8(5):142.
doi pubmed - Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720-741.
pubmed - Taylor DN, Blaser MJ. The epidemiology of Helicobacter pylori infection. Epidemiol Rev. 1991;13:42-59.
doi - Dorer MS, Talarico S, Salama NR. Helicobacter pylori’s unconventional role in health and disease. PLoS Pathog. 2009;5(10):e1000544.
doi pubmed - Kuipers EJ, Perez-Perez GI, Meuwissen SG, Blaser MJ. Helicobacter pylori and atrophic gastritis: importance of the cagA status. J Natl Cancer Inst. 1995;87(23):1777-1780.
doi pubmed - Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A. 1996;93(25):14648-14653.
doi pubmed - Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111-2115.
pubmed - Perng CL, Lin HJ, Sun IC, Tseng GY, Facg. Helicobacter pylori cagA, iceA and vacA status in Taiwanese patients with peptic ulcer and gastritis. J Gastroenterol Hepatol. 2003;18(11):1244-1249.
doi pubmed - Wu J, Xu S, Zhu Y. Helicobacter pylori CagA: a critical destroyer of the gastric epithelial barrier. Dig Dis Sci. 2013;58(7):1830-1837.
doi pubmed - Atherton JC, Cao P, Peek RM, Jr., Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):17771-17777.
doi pubmed - Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol. 2005;3(4):320-332.
doi pubmed - van Doorn LJ, Schneeberger PM, Nouhan N, Plaisier AP, Quint WG, de Boer WA. Importance of Helicobacter pylori cagA and vacA status for the efficacy of antibiotic treatment. Gut. 2000;46(3):321-326.
doi pubmed - Dabiri H, Bolfion M, Mirsalehian A, Rezadehbashi M, Jafari F, Shokrzadeh L, Sahebekhtiari N, et al. Analysis of Helicobacter pylori genotypes in Afghani and Iranian isolates. Pol J Microbiol. 2010;59(1):61-66.
pubmed - Abu-Lubad M, et al. Molecular Epidemiology of Helicobacter pylori in Dental Plaque among Jordanians; A probable source for infection and treatment failure. J Pure Appl Microbiol. 2017;11(1):9-14.
doi - Parsonnet J. The incidence of Helicobacter pylori infection. Aliment Pharmacol Ther. 1995;9(Suppl 2):45-51.
pubmed - Pounder RE, Ng D. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol Ther. 1995;9(Suppl 2):33-39.
pubmed - Amiri N, Abiri R, Eyvazi M, Zolfaghari MR, Alvandi A. The frequency of Helicobacter pylori in dental plaque is possibly underestimated. Arch Oral Biol. 2015;60(5):782-788.
doi pubmed - Momtaz H, Souod N, Dabiri H, Sarshar M. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World J Gastroenterol. 2012;18(17):2105-2111.
doi pubmed - Berroteran A, Perrone M, Correnti M, Cavazza ME, Tombazzi C, Goncalvez R, Lecuna V. Detection of Helicobacter pylori DNA in the oral cavity and gastroduodenal system of a Venezuelan population. J Med Microbiol. 2002;51(9):764-770.
doi pubmed - Medina ML, Medina MG, Martin GT, Picon SO, Bancalari A, Merino LA. Molecular detection of Helicobacter pylori in oral samples from patients suffering digestive pathologies. Med Oral Patol Oral Cir Bucal. 2010;15(1):e38-42.
pubmed - Souod N, Kargar M, Doosti A, Ranjbar R, Sarshar M. Genetic analysis of cagA and vacA genes in helicobacter pylori isolates and their relationship with gastroduodenal diseases in the west of Iran. Iran Red Crescent Med J. 2013;15(5):371-375.
doi pubmed - Ben Mansour K, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, Ben Mami N, et al. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann Clin Microbiol Antimicrob. 2010;9:10.
doi pubmed - Hatakeyama M, Brzozowski T. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2006;11(Suppl 1):14-20.
doi pubmed - Sicinschi LA, Correa P, Bravo LE, Schneider BG. Detection and typing of Helicobacter pylori cagA/vacA genes by radioactive, one-step polymerase chain reaction in stool samples from children. J Microbiol Methods. 2003;52(2):197-207.
doi - Rasmussen LT, Labio RW, Gatti LL, Silva LC, Queiroz VF, Smith Mde A, Payao SL. Helicobacter pylori detection in gastric biopsies, saliva and dental plaque of Brazilian dyspeptic patients. Mem Inst Oswaldo Cruz. 2010;105(3):326-330.
doi pubmed - Nimri LF, Matalka I, Bani Hani K, Ibrahim M. Helicobacter pylori genotypes identified in gastric biopsy specimens from Jordanian patients. BMC Gastroenterol. 2006;6:27.
doi pubmed - Silva DG, Stevens RH, Macedo JM, Albano RM, Falabella ME, Veerman EC, Tinoco EM. Detection of cytotoxin genotypes of Helicobacter pylori in stomach, saliva and dental plaque. Arch Oral Biol. 2009;54(7):684-688.
doi pubmed - Allaker RP, Young KA, Hardie JM, Domizio P, Meadows NJ. Prevalence of helicobacter pylori at oral and gastrointestinal sites in children: evidence for possible oral-to-oral transmission. J Med Microbiol. 2002;51(4):312-317.
doi pubmed - Faundez G, Troncoso M, Figueroa G. cagA and vacA in strains of Helicobacter pylori from ulcer and non-ulcerative dyspepsia patients. BMC Gastroenterol. 2002;2:20.
doi pubmed - Perez-Perez GI, Peek RM, Jr., Atherton JC, Blaser MJ, Cover TL. Detection of anti-VacA antibody responses in serum and gastric juice samples using type s1/m1 and s2/m2 Helicobacter pylori VacA antigens. Clin Diagn Lab Immunol. 1999;6(4):489-493.
pubmed - Torres LE, Melian K, Moreno A, Alonso J, Sabatier CA, Hernandez M, Bermudez L, et al. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World J Gastroenterol. 2009;15(2):204-210.
doi pubmed - Prinz C, Schwendy S, Voland P. H pylori and gastric cancer: shifting the global burden. World J Gastroenterol. 2006;12(34):5458-5464.
doi pubmed - Assumpcao MB, Martins LC, Melo Barbosa HP, Barile KA, de Almeida SS, Assumpcao PP, Corvelo TC. Helicobacter pylori in dental plaque and stomach of patients from Northern Brazil. World J Gastroenterol. 2010;16(24):3033-3039.
doi pubmed - Roman-Roman A, Giono-Cerezo S, Camorlinga-Ponce M, Martinez-Carrillo DN, Loaiza-Loeza S, Fernandez-Tilapa G. vacA genotypes of Helicobacter pylori in the oral cavity and stomach of patients with chronic gastritis and gastric ulcer. Enferm Infecc Microbiol Clin. 2013;31(3):130-135.
doi pubmed - Khandaker K, Palmer KR, Eastwood MA, Scott AC, Desai M, Owen RJ. DNA fingerprints of Helicobacter pylori from mouth and antrum of patients with chronic ulcer dyspepsia. Lancet. 1993;342(8873):751.
doi - Shames B, Krajden S, Fuksa M, Babida C, Penner JL. Evidence for the occurrence of the same strain of Campylobacter pylori in the stomach and dental plaque. J Clin Microbiol. 1989;27(12):2849-2850.
pubmed - Ferguson DA, Jr., Li C, Patel NR, Mayberry WR, Chi DS, Thomas E. Isolation of Helicobacter pylori from saliva. J Clin Microbiol. 1993;31(10):2802-2804.
pubmed - Cellini L, Allocati N, Piattelli A, Petrelli I, Fanci P, Dainelli B. Microbiological evidence of Helicobacter pylori from dental plaque in dyspeptic patients. New Microbiol. 1995;18(2):187-192.
pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.


