| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website http://www.gastrores.org |
Original Article
Volume 4, Number 6, December 2011, pages 262-267
Helicobacter Pylori DNA in Liver Tissues From Chronic Hepatitis C Egyptian Patients
Moushira A. Mahmouda, c, Loaa A. Tag Eldena, Mohamed M. Awadb, Henock A. Hailea
aDepartment of Medical Biochemistry, Suez Canal Faculty of medicine, Ismailia, Egypt
bDepartment of Internal Medicine, Suez Canal Faculty of medicine, Ismailia, Egypt
cCorresponding author: Moushira A. Mahmoud, Department of Biochemistry, Faculty of Medicine, Suez Canal University, Ismailia, Egypt
Manuscript accepted for publication September 26, 2011
Short title: H. pylori DNA in Liver From CHC Egyptian Patients
doi: https://doi.org/10.4021/gr356w
| Abstract | ▴Top |
Background: Hepatitis C virus (HCV) is considered the most common etiology of chronic liver disease in Egypt, which may progress to cirrhosis and hepatocellular carcinoma (HCC). Previous studies have documented an association between Helicobacter pylori (H. pylori) infection and liver cirrhosis with or without HCC. This study aimed to investigate the presence of H. pylori DNA in the liver tissue of Egyptian patients with chronic hepatitis C (CHC).
Methods: Fifty-two CHC Egyptian patients were enrolled in this study. Plasma anti-H. pylori IgG was assessed with ELISA. Liver biopsies were tested for presence of Helicobacter DNA using genus specific nested polymerase chain reaction (PCR) and species was identified by sequencing.
Results: Anti-H. pylori IgG was detected in 31/52 (59.6%) CHC patients while Helicobacter DNA was detected in 6 (11.5%) patients, all were H. Pylori by sequencing. Helicobacter DNA was more frequent in patients with high stage liver fibrosis (33.3%) than in those with low stage fibrosis (2.7%) (P = 0.006). There was no association between the presence of H. pylori DNA in the liver and age, gender of patients, liver function tests, AFP levels or viral load.
Conclusions: These data confirm the presence of H. pylori DNA in liver of some CHC Egyptian patients and suggest an association of this bacterium with progression of liver fibrosis.
Keywords: HCV; Chronic hepatitis C; H. pylori; Liver fibrosis
| Introduction | ▴Top |
Hepatitis C Virus (HCV) infects an estimated 170 million persons worldwide. The prevalence of HCV infection varies throughout the world, with the highest prevalence (14.7%) reported in Egypt [1]. More than 70% of HCV-infected individuals develop chronic disease, which can progress to liver cirrhosis and hepatocellular carcinoma (HCC). The course of HCV related hepatic disease varies markedly from one patient to another. It is affected by age at exposure, duration of infection, alcohol intake, male gender, viral immune response [2]. However, even in the absence of these factors, disease progression may be observed in some patients, suggesting the role of other factors which remain to be identified. Host genetic factors or environmental factors, such as a bacterial co-infection, could be involved [3].
H. pylori is a Gram-negative organism that colonizes gastric mucosa and known to cause chronic gastritis, peptic ulcers, and gastric adenocarcinoma [4]. However, several separate research groups have detected Helicobacter spp. in liver tissue of patients with different hepato-biliary diseases. Nilsson et al have identified H. pylori in human liver samples from patients suffering from primary sclerosing cholangitis and primary biliary cirrhosis [5]. Tolia et al have demonstrated, by PCR, the presence of genomic sequences of Helicobacter spp. in the liver tissue of 40 patients with miscellaneous liver diseases; a further analysis by sequencing revealed that most of these species were H. pylori [6]. Several researchers reported that H. Pylori DNA can be found in patients with primary liver carcinoma and probably linked to the carcinogenic process in the liver [7, 8]. Moreover, studies from different countries documented the detection of H. pylori DNA in the liver tissue of patients with HCV-related chronic hepatitis, cirrhosis and HCC, suggesting that these bacteria could be implicated in the progression of CHC to cirrhosis and HCC [9-11].
In Egypt, a recent study demonstrated that the seroprevalence of H. pylori increased significantly in the HCV-infected patients when compared to healthy controls. Moreover, the researchers found that the prevalence of H. pylori infection increased significantly from chronic active hepatitis to cirrhosis [12]. However, up to our knowledge, the presence of H. pylori in the liver tissue of HCV Egyptian patients was not previously investigated. Therefore, this study was conducted to investigate the presence of H. pylori DNA in liver biopsies from Egyptian patients with CHC.
| Patients and Methods | ▴Top |
Fifty two consecutive patients with CHC were included in the study. Those patients were required to have a percutaneous liver biopsy at the Gastroenterology and Hepatology Unit of Suez Canal University Hospital, as a line of their management. All were positive for anti-HCV and HCV RNA. Chronic hepatitis C was diagnosed by having either elevated or fluctuating ALT levels for more than 6 months and/or bright liver appearance on abdominal ultrasonography [13]. Patients with other causes of liver disease, including hepatitis B, were excluded from the study. The study was approved by the Research Ethics Committee of the Suez Canal Faculty of Medicine and informed consents were obtained from all study subjects. Liver function tests, AFP and anti-schistosomal antibodies were measured using commercially available kits. The HCV viral load was quantified using Real Time PCR technique in an ABI PRISM® 7000 thermocycler (Applied Biosystems, Foster City, CA).
Liver tissues processing
Each liver biopsy sample was divided into two parts: One was fixed in formalin and embedded in paraffin wax for conventional histological evaluation; the other was immediately frozen and stored at -80 °C until further molecular analysis. The formalin-fixed specimens were examined by pathologists at pathology department of Suez Canal University Hospital. Liver fibrosis was staged on a 0 - 6 scale according to Ishak score [14]. Patients were divided into two groups based on fibrosis score: “low stage” [F0-F3] and “high stage” [F4-F6] liver fibrosis.
Detection of anti-H. pylori IgG
Plasma samples were tested for anti-H. pylori IgG antibody using a commercial test kit, AccuBind™ ELISA Microwells (Monobind Inc, Lake Forest, USA), according to the manufacturer’s instruction. Results were considered positive when higher than 20 U/mL.
PCR amplification with Helicobacter genus-specific primers
DNA was extracted from frozen liver tissues using Wizard®SV Genomic DNA Purification System (Promega Corporation, Madison, USA). DNA quantitation was performed using the NanoDrop® (ND)-1000 Spectrophotometer (NanoDrop Technologies Inc., Washington,USA). The extracted DNA was stored at -20 °C until further used. Nested PCR was performed with genus-specific primers for the Helicobacter 16S ribosomal RNA gene (16S rDNA). The primers (Helinest-S & R, Heli-S & R) used in our study were reported to amplify 26 species of Helicobacter genus [15].
First amplification
Amplification was carried out in a total volume of 50 µL containing 1 µg DNA, 25 µL DreamTaq™ Green PCR Master Mix (2 x) (Fermentas, CA, USA) and 50 pM of each primer (Helinest-S 5’-ATTAGTGGCGCACGGGTGAGTAA-3’ and Helinest-R 5’-TTTAGCATCCCGACTTAAGGC-3’). The reaction mixture was initially denatured for 2 minutes at 94 °C then amplified for 35 cycles as follows: denaturation for 30 seconds at 94 °C, primer annealing for 30 seconds at 55 °C and extension for 1.5 minutes at 72°C. A final extension step was done for 5 minutes at 72 °C (Thermocycler Robocycler Gradiant 96-STRATAGEN®, LA, USA).
Second amplification
Five microliter of amplicon from the first amplification step was used with primers Heli-S (5’-GAACCTTACCTAGGCTTGACATTG-3’) and Heli-R (5’-GGTGAGTACAAGACCCGGGAA-3’) and amplification was repeated as in the first amplification. PCR amplicons were visualised on a 2% agarose gel stained with ethidium bromide. The expected product size of the amplicon was 480 bp.
DNA sequencing
PCR products were sequenced as described previously [15]. Sequence homology searches of the PCR products were performed with the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information).
Statistical analysis
Data were analyzed using Statistical Package for the Social Sciences, Version 10 (SPSS Inc. Chicago, IL, USA). Different characteristics of study participants were described using mean ± SD, median (range) or number (percentage) as appropriate. Continuous variables were compared using the Student’s unpaired t-test or Mann-Whitney test. The association between categorical parameters was performed using the Chi-square test or Fisher’s exact test. P-values < 0.05 were considered statistically significant.
| Results | ▴Top |
Table 1 summarizes the main demographic and laboratory characteristics of the 52 CHC patients. The mean age of patients was 42.5 ± 7.4 and only 5 were positive for anti-Schistosomal antibodies.
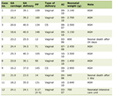 Click to view | Table 1. Characteristics of the Chronic Hepatitis C Patients |
Anti-H. pylori IgG was detected in the plasma of 31/52 (59.6%) patients. There were no significant differences in the demographic and laboratory characteristics among anti-H. pylori IgG-positive and anti-H. pylori IgG-negative patients (Table 2).
 Click to view | Table 2. Comparison Between Anti-H. pylori IgG Positive and Negative Patients Regarding Demographic and Laboratory Data |
Helicobacter DNA was present in 6/52 (11.5%) liver biopsy samples, collected from CHC patients, using Helicobacter genus specific 16 S rRNA gene primers (Fig. 1). The PCR products were sequenced and H. pylori like organisms were identified. All cases positive for H. pylori DNA in the tissue samples were positive for plasma anti-H. pylori IgG and negative for anti-schistosomal antibodies.
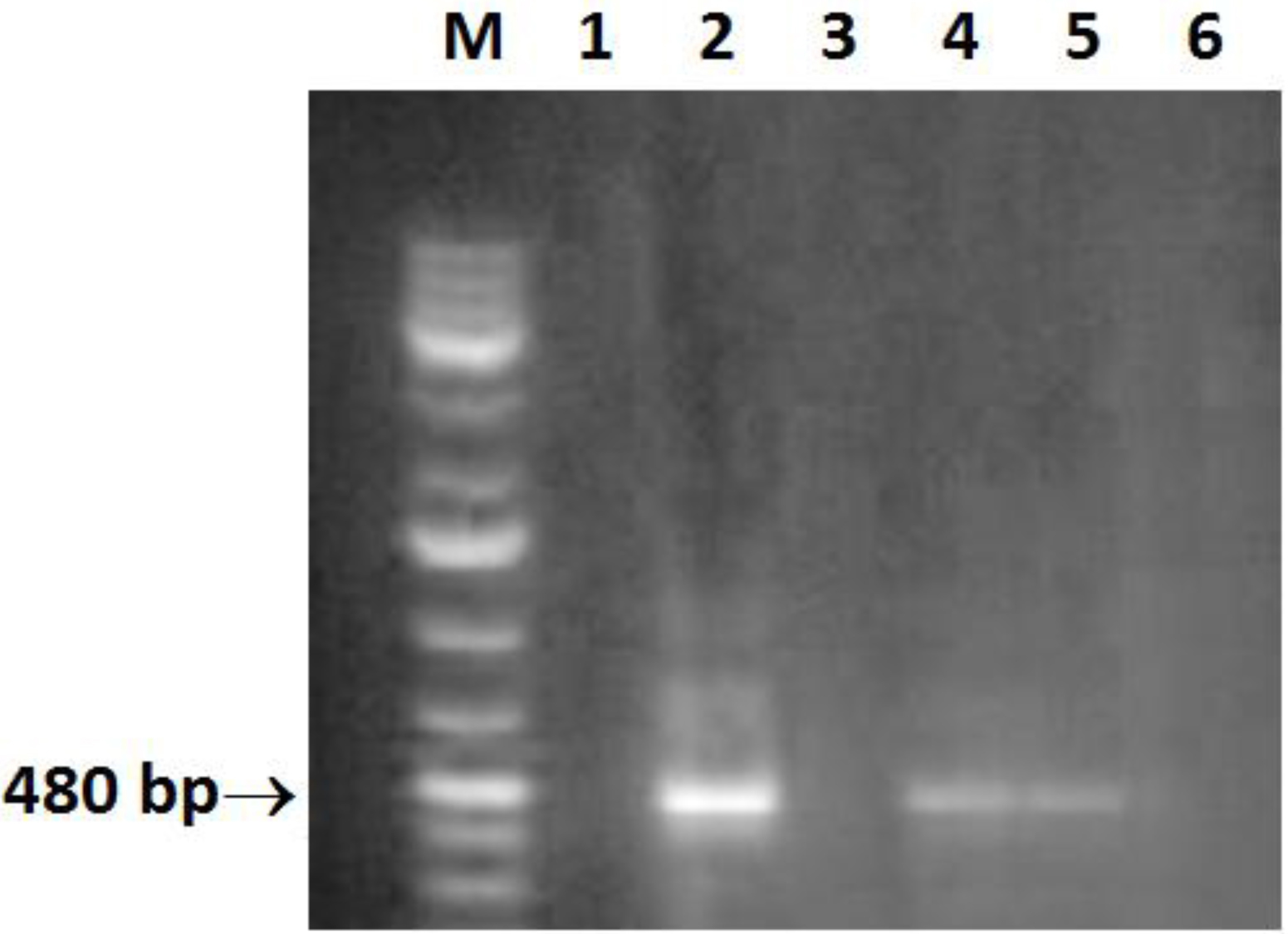 Click for large image | Figure 1. Amplification of a 480-bp 16S rRNA DNA fragment of Helicobacter. Lane M: molecular size marker (100 - 1000 bp); lane 2, 4, 5: positive samples; lane 6: negative control (double-distilled water). |
Helicobacter DNA positive cases were more frequent in CHC patients with high stage liver fibrosis 5/15(33.3%) than in low stage liver fibrosis 1/37 (2.7%). Thus, there was statistically significant (P = 0.0057) association between presence of H. Pylori DNA in the liver and stage of fibrosis. However, there was no association between the presence of H. Pylori DNA in the liver of CHC patients and age, gender, liver function tests, AFP levels or HCV viral load (Table 3).
 Click to view | Table 3. Comparison Between H. pylori DNA Positive and Negative Patients Regarding Demographic and Laboratory Data |
| Discussion | ▴Top |
Since the discovery of presence of Helicobacter species DNA in liver material from patients with liver disease, several studies were conducted to determine the role of these bacteria in the evolution of hepatic lesions to cirrhosis and HCC. Determinants of this evolution are not yet fully understood, including those occurring in HCV positive patients [10].
In the present study, H. Pylori DNA was detected in liver biopsies of 11.5% Egyptian patients with CHC which agrees with previous studies [10, 11, 16]. H. pylori DNA positive cases tended to be higher in CHC patients with high stage liver fibrosis (33.3%) than in low stage liver fibrosis (2.7%) (P = 0.0057). Our finding seems similar to the study of Castera et al in which they demonstrated higher prevalence of Helicobacter DNA in the liver samples from patients with hepatitis C cirrhosis than in those from HCV-infected patients without cirrhosis or from controls [16]. It has been suggested that H. pylori infection may affect the clearance of a concomitant viral infection through down regulation of the virus-specific T-lymphocyte helper-1 cytokine and T-lymphocyte suppressor responses [17]. Moreover, H. pylori DNA has been detected in the liver tissue of patients with cirrhosis and more frequently in HCC [10, 18]. The presence of H. pylori DNA at a higher frequency in the liver of patients with high stage fibrosis than low stage fibrosis may suggest the participation of this bacterium in the progress of the chronic hepatitis to HCC.
The fact that the DNA sequence, obtained from positive PCR of Helicobacter spp. specific 16 S rRNA gene, was analogous to H. pylori encourages the speculation that the presence of Helicobacter DNA in human liver tissue might reflect the transport of H. pylori of gastric origin or its DNA to the liver [15]. Verhoef et al reported that gastric colonization with a specific subset of Helicobacter strains is associated with the induction of HCC, either directly via colonization of the liver or indirectly via secretion of specific toxins by Helicobacter residing in the stomach [19]. Recently, oral administration of H. pylori to C57BL/6 mice, for two years, resulted in not only gastric lesions but also liver lesions, including inflammation, cirrhosis and hepatocyte hyperplasia with atypia. Thus, H. pylori inoculated orally could reach the liver and cause inflammation as an independent etiological factor [20].
In the present study, H. pylori DNA was found only in liver tissue from anti-H. pylori IgG positive patients that reflects stomach inoculation with H. pylori in these patients. This observation comes in agreement with several previous studies in which all helicobacter DNA positive liver samples were seropositive for H. pylori IgG [11, 16, 21]. The bacterium may pass from the stomach to the liver through the duodenum and biliary tract, or may arrive in the liver from the circulation through the hepatic portal vein [22]. Huang et al found H. pylori DNA in the peripheral blood of patients with duodenal ulcers infected with H. pylori, which suggested that H. pylori may spread to the liver by the blood-borne route [23].
This study also revealed the absence of any significant difference in liver function tests (ALT, AST, albumin, and bilirubin), AFP levels or HCV viral load among H. pylori DNA positive and H. pylori DNA negative patients. The lack of difference in liver function tests among these groups of patients indicates that the Helicobacter positivity may not primarily relate to severe liver damage. Our findings showed similarity with previous studies which, reported absence of association of any of these factors with respect to presence of H. pylori DNA in the liver of CHC and other chronic liver diseases patients [16, 24].
In conclusion, H. pylori DNA could be detected in liver of some CHC Egyptian patients and its presence is associated with more advanced liver fibrosis. Further studies are needed to investigate the role of H. pylori in the progression of chronic liver diseases to cirrhosis and its role in the development of HCC.
| References | ▴Top |
- El-Zanaty F, Way A. Egypt Demographic and Health Survey 2008. Egyptian Ministry of Health (El-Zanaty and Associates and Macro International, Cairo) 2009; pp 431.
- Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3(2):47-52.
pubmed - de Magalhaes Queiroz DM, Santos A. Isolation of a Helicobacter strain from the human liver. Gastroenterology. 2001;121(4):1023-1024.
pubmed - Blaser MJ. Helicobacter pylori and gastric diseases. BMJ. 1998;316(7143):1507-1510.
pubmed doi - Nilsson HO, Taneera J, Castedal M, Glatz E, Olsson R, Wadstrom T. Identification of Helicobacter pylori and other Helicobacter species by PCR, hybridization, and partial DNA sequencing in human liver samples from patients with primary sclerosing cholangitis or primary biliary cirrhosis. J Clin Microbiol. 2000;38(3):1072-1076.
pubmed - Tolia V, Nilsson HO, Boyer K, Wuerth A, Al-Soud WA, Rabah R, Wadstrom T. Detection of Helicobacter ganmani-like 16S rDNA in pediatric liver tissue. Helicobacter. 2004;9(5):460-468.
pubmed doi - Huang Y, Fan XG, Wang ZM, Zhou JH, Tian XF, Li N. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J Clin Pathol. 2004;57(12):1273-1277.
pubmed doi - Xuan SY, Li N, Qiang X, Zhou RR, Shi YX, Jiang WJ. Helicobacter infection in hepatocellular carcinoma tissue. World J Gastroenterol. 2006;12(15):2335-2340.
pubmed - Ponzetto A, Pellicano R, Leone N, Cutufia MA, Turrini F, Grigioni WF, D'Errico A,
et al . Helicobacter infection and cirrhosis in hepatitis C virus carriage: is it an innocent bystander or a troublemaker?. Med Hypotheses. 2000;54(2):275-277.
pubmed doi - Rocha M, Avenaud P, Menard A, Le Bail B, Balabaud C, Bioulac-Sage P, de Magalhaes Queiroz DM,
et al . Association of Helicobacter species with hepatitis C cirrhosis with or without hepatocellular carcinoma. Gut. 2005;54(3):396-401.
pubmed doi - Dore MP, Realdi G, Mura D, Graham DY, Sepulveda AR. Helicobacter infection in patients with HCV-related chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Dig Dis Sci. 2002;47(7):1638-1643.
pubmed doi - El-Masry S, El-Shahat M, Badra G, Aboel-Nour MF, Lotfy M. Helicobacter pylori and Hepatitis C Virus Coinfection in Egyptian Patients. J Glob Infect Dis. 2010;2(1):4-9.
pubmed doi - Joseph AE, Dewbury KC, McGuire PG. Ultrasound in the detection of chronic liver disease (the "bright liver"). Br J Radiol. 1979;52(615):184-188.
pubmed doi - Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H,
et al . Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696-699.
pubmed doi - Pellicano R, Mazzaferro V, Grigioni WF, Cutufia MA, Fagoonee S, Silengo L, Rizzetto M,
et al . Helicobacter species sequences in liver samples from patients with and without hepatocellular carcinoma. World J Gastroenterol. 2004;10(4):598-601.
pubmed - Castera L, Pedeboscq A, Rocha M, Le Bail B, Asencio C, de Ledinghen V, Bernard PH,
et al . Relationship between the severity of hepatitis C virus-related liver disease and the presence of Helicobacter species in the liver: a prospective study. World J Gastroenterol. 2006;12(45):7278-7284.
pubmed - Shirai M, Arichi T, Nakazawa T, Berzofsky JA. Persistent infection by Helicobacter pylori down-modulates virus-specific CD8+ cytotoxic T cell response and prolongs viral infection. J Infect Dis. 1998;177(1):72-80.
pubmed doi - Chen R, Chen XP, Lin QQ, Lin BL, Cao HJ. Detection of Helicobacter species related genes coding for 16S rRNA in the liver tissue of patients with chronic liver disease. Nan Fang Yi Ke Da Xue Xue Bao. 2010;30(1):131-132.
pubmed - Verhoef C, Pot RG, de Man RA, Zondervan PE, Kuipers EJ, JN IJ, Kusters JG. Detection of identical Helicobacter DNA in the stomach and in the non-cirrhotic liver of patients with hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 2003;15(11):1171-1174.
pubmed doi - Tian XF, Fan XG, Huang X, Fu CY, Dai H, Huang Y. A two-year animal experimental study on the pathological effects of Helicobacter pylori on liver tissues. Zhonghua Gan Zang Bing Za Zhi. 2008;16(2):129-133.
pubmed - Stalke P, Al-Soud WA, Bielawski KP, Bakowska A, Trocha H, Stepinski J, Wadstrom T. Detection of Helicobacter species in liver and stomach tissues of patients with chronic liver diseases using polymerase chain reaction-denaturing gradient gel electrophoresis and immunohistochemistry. Scand J Gastroenterol. 2005;40(9):1032-1041.
pubmed doi - Pellicano R, Menard A, Rizzetto M, Megraud F. Helicobacter species and liver diseases: association or causation?. Lancet Infect Dis. 2008;8(4):254-260.
pubmed doi - Huang Y, Fan XG, Tang ZS, Liu L, Tian XF, Li N. Detection of Helicobacter pylori DNA in peripheral blood from patients with peptic ulcer or gastritis. APMIS. 2006;114(12):851-856.
pubmed doi - Petrenkienë V, Vitkauskienë A, Jonaitis L, Kupèinskas L, Wadström T. Detection of Helicobacter spp. in liver biopsy specimens. Acta Medica Lituanica. 2004;11(3):31-35.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.


