| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website http://www.gastrores.org |
Original Article
Volume 3, Number 3, June 2010, pages 101-105
A RecQ Protein-like 5 Haplotype is Associated With Colon Cancer
Heather M. Ochs-Balcoma, b, Cheryl L.Thompsonb, c, d, Sarah Plummere, Guangbin Luof, Thomas C. Tuckerg, Graham Caseye, Li Lib, c, d, h
aDepartment of Social and Preventive Medicine, State University of New York at Buffalo, Buffalo, New York, 14214, USA
bDepartment of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland, Ohio, 44106, USA
cDepartment of Family Medicine, Case Western Reserve University/University Hospitals Case Medical Center, Cleveland, Ohio, 44106, USA
dCase Center for Transdisciplinary Research on Energetics and Cancer, Case Comprehensive Cancer Center, Case Western Reserve University, Cleveland, Ohio, 44106, USA
eDepartment of Preventive Medicine, University of Southern California, Los Angeles, California, 90089, USA
fDepartment of Genetics, Case Western Reserve University, 44106, Cleveland, Ohio, USA
gCancer Control Program, University of Kentucky, Lexington, Kentucky, 40536, USA
hCorresponding author: Department of Family Medicine, Case Comprehensive Cancer Center, Case Western Reserve University, 11001 Cedar Ave, Suite 306, Cleveland, OH 44106, USA;
Manuscript accepted for publication June 18, 2010
Short title: RECQL5 and Colon Cancer
doi: https://doi.org/10.4021/gr2010.06.214w
| Abstract | ▴Top |
Background: Emerging murine model data suggests RecQ protein-like 5 (RECQL5) is a tumor suppressor gene. The goal of our study was to test whether RECQL5 gene variants are associated with colon cancer susceptibility.
Methods: We examined the association of two haplotype-tagging SNPs in RECQL5 and colon cancer in a population-based study of 390 colon cancer cases and 464 population controls.
Results: While both crude and covariate-adjusted single SNP analyses were only suggestive for an association with borderline significance (p = 0.07), haplotype analysis shows that individuals carrying the T-G haplotype (rs820196 common allele and rs4789223 minor allele) were at significantly increased risk for colon cancer (OR = 1.34, 95% CI = 1.02-1.76, p = 0.05). Adjustment for age, sex, body mass index, non-steroidal anti-inflammatory use and family history of colon cancer did not alter the results.
Conclusions: These results suggest that a haplotype harboring the minor allele of rs4789223 is associated with colon cancer risk. Further study of RECQL5 as a colon cancer susceptibility gene is warranted, particularly with respect to variants in linkage disequilibrium with rs4789223.
Keywords: RECQL5; Colon cancer; Haplotype; Single Nucleotide Polymorphism
| Introduction | ▴Top |
RECQL5 is a member of the RecQ helicase family which includes a number of disease-causing genes implicated in several cancer prone syndrome such as Bloom syndrome, Werner syndrome, and Rothmund-Thomson syndrome [1]. Recent work in Apcmin/+ mice harboring Recql5 deficiency (Recql5-/-) showed marked increase in microadenoma formation compared to Recql5+/+ mice [2]. Another recent study showed that mice deficient for Recql5, the mouse homologue of human RECQL5 are predisposed to a wide variety of other sporadic cancers, establishing this protein as an important tumor suppressor in mice [3]. This evidence along with the fact that mouse Recql5 and human RECQL5 are highly conserved [3], suggests human RECQL5 may also have an important tumor suppressing function and therefore certain genetic variants that affect the function of this RECQL5 may then alter the risk of cancer in humans. In this study, we tested the hypothesis that RECQL5 variants are associated with colon cancer in a population-based case control study.
| Patients and Methods | ▴Top |
Study population
The study design and methodology for this population-based case-control study has been described elsewhere, and all human subjects approvals were obtained [4]. Briefly, eligible cases were identified through the population-based Kentucky Cancer Registry which is part of both the National Cancer Institutes' Surveillance, Epidemiology and End Results (SEER) program and the Centers for Disease Control and Preventions' National Program of Cancer Registries (NPCR). Residents living in Kentucky at the time of diagnosis between 2003 and 2006 were included in this study. We identified all incident and primary colon cancer cases reported within 6 months of diagnosis. We used random digital dialing to recruit controls. The area codes and exchanges of the cases were used as a proxy for frequency match of the residential locations of the cases, along with randomly-generated 4 digit numbers for recruitment of cancer-free controls. We collected information on demographics, family history of colorectal cancer, and personal cancer history via telephone questionnaire. Each participant received a phlebotomy kit with instructions for fasting blood sample collection, consent forms, and a self-administered lifestyle risk factor questionnaire (RFQ) (http://epi.grants.cancer.gov/documents/CFR/center_questionnaires/Colon/LA/ColonRiskFactor_USC.pdf) developed by the National Cancer Institute Colon Cancer Familial Cancer Registry to collect detailed information on family history of colorectal cancer, lifestyle and behavioral risk factors. Individuals with known inflammatory bowel diseases, family history of familial adenomatous polyposis, and hereditary non-polyposis colorectal cancer were excluded from the study, and controls were required to be at least 30 years of age. All participants provided written informed consent. The study was approved by the Institutional Review Boards of the University of Kentucky, Lexington, and Case Western Reserve University/University Hospitals of Cleveland. The participation rates were 72.2% for the cases, and 62.5% for eligible controls.
SNP selection and genotyping
Common genetic variation in the RECQL5 gene was identified by querying the Genome Variation Server (http://gvs.gs.washington.edu/GVS) for SNPs within RECQL5 and 5 kb upstream and downstream with a minor allele frequency (MAF) of at least 5%. Haplotype tagging SNPs were selected based on MAF and linkage disequilibrium (LD) patterns in the HapMap Caucasian samples with preference given to non-synonymous coding variants. Caucasian samples were used as a reference as over 90% of our sample self-report as Caucasian. A single non-synonymous coding SNP was identified (rs820196) in this region. No other non-synonymous coding or regulatory SNPs were identified. The addition of one more SNP (rs4789223) was identified to sufficiently tag the entire region with 85% coverage.
Blood samples were shipped overnight on frozen ice pack. Upon receipt, samples were spun for 15 minutes at 600 x g and aliquots of plasma and concentrated buffy coat were prepared and frozen at -80°C. Genomic DNA was extracted from thawed buffy coat fractions using the Qiagen EZ1 Biorobot. Genotypes were determined using 1.25 ng of genomic DNA with predesigned Taqman assays from Applied Biosystems (C_7978409_10, and C_7978435_20) and RealMasterMixProbe +ROX (5 Prime). Assays were read using the 7900HT Real-Time PCR system (Applied Biosystems). Two percent of samples were repeated with a concordance of 100%. Neither SNP deviated significantly from Hardy-Weinberg proportions.
Statistical analysis
We limited our analyses to Caucasians only due to the few individuals in other racial/ethnic groups (N = 61). Of 397 cases and 467 controls that completed the study, seven cases and three controls were excluded due to missing SNP data. The final set for analysis includes 390 cases and 464 controls.
We examined differences in demographic and risk factors using chi-square tests and student’s t-tests. We estimated allele and genotype frequencies and assessed deviations from Hardy-Weinberg equilibrium in cases and controls separately using chi-square tests with 1 degree of freedom. We computed odds ratios (ORs) and 95% confidence intervals using unconditional logistic regression models under log-additive genetic models for each of SNP. We estimated both crude ORs and ORs adjusted for age, sex, non-steroidal anti-inflammatory drug (NSAID) use, body mass index (BMI), and any family history of colorectal cancer. NSAID use was defined as self-reported intake of aspirin or ibuprofen at least twice weekly for a period of six months or longer.
We estimated haplotype frequencies of 2-SNP haplotypes using the expectation maximization algorithm within the UNPHASED software, and estimated risk associated with each haplotype with the most common haplotype as the reference category using unconditional logistic regression [5, 6].
| Results | ▴Top |
We selected two tagSNPs to represent genetic variation at the RECQL5 locus. The rs820196 SNP is located in exon 1 (71139134 in build 36) and is a nonsynonymous mutation that results in an amino acid change from aspartic acid to glycine. The rs4789223 SNP is intronic.
Table 1 shows that similar to previous reports on this sample [4], cases were more likely have a higher BMI (p = 0.001) and a family history of colorectal cancer (p = 0.001). Cases were also slightly older at diagnosis than the controls at time of recruitment; the mean age was 63.0 and 58.1 years for cases and controls, respectively. The cases comprised a higher proportion of males than the controls (p = 0.001).
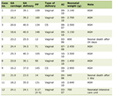 Click to view | Table 1. Kentucky Colon Cancer Study Demographics |
Table 2 shows the allele and genotype frequencies for RECQL5 SNPs. The minor allele frequency for rs820196 in our control sample (0.23) was slightly higher than the HapMap CEPH data (0.21). The MAF for rs4789223 was very similar; our control frequency was 0.39 and Hapmap was 0.38. Allelic association tests were not significant for rs4789223 (p = 0.07) or rs820196 (p = 0.60). Table 2 shows the adjusted OR of 1.21 (95% CI = 0.98 -1.49).
 Click to view | Table 2. Allele and Genotype Frequencies and Association Tests for RECQL5 SNPs |
The haplotype results in Table 3 suggest that individuals who carry the T-G haplotype, that is the common allele of rs820196 and the minor allele of rs4709223, are at increased risk compared to individuals who carry the common alleles together on a haplotype, crude OR: 1.34, 95% CI = 1.02 - 1.76. The T-G haplotype was more common in cases; approximately 20% of the cases and 16% of the controls carried this haplotype. Adjustment for covariates only strengthened the estimates.
 Click to view | Table 3. RECQL5 Colon Cancer Haplotype Association Tests |
| Discussion | ▴Top |
Colon cancer is a complex disease that is in part defined by the involvement of multiple genes with small effects influencing susceptibility. The search for susceptibility genes for colon cancer remains an active area of research in the form of both genome-wide association studies (GWAS) and candidate gene studies, each of which have their respective strengths and limitations for gene discovery. Many independent associations have been identified using the genome-wide approach, and these regions do not include the region on chr17 harboring RECQL5 [7]. Nonetheless, many other important candidate genes are likely to remain undiscovered using the GWAS approach, particularly because the large number of statistical tests are prohibitive to examining more than single SNP models (i.e. multi-allelic or haplotype models) at the first stage analysis. We hypothesized that RECQL5 is a novel candidate gene for colon cancer given the known involvement in DNA repair and homologous recombination and association of other members of this family with known cancer disorders. Our tag-SNP association analysis did not reveal evidence for an association with colon cancer and RECQL5. We did however find a significant association with a haplotype harboring the minor allele of rs4789223. These results are not surprising in a complex disease such as colon cancer, where multiple genes may have small effects.
To date, there are no known diseases or syndromes associated with variants or mutations in RECQL5. First cloned in silico in 1998, RECQL5 was found to be expressed in most tissues, especially pancreas and testis, unlike other members of the family whose expression appears to be more tissue-specific [8]. RECQL5 is known to be alternatively spliced to produce three different isoforms, RECQL5α, RECQL5β, and RECQL5γ [9, 10].
The rs820196 SNP in RECQL5 is a nonsynonymous coding SNP in exon 9, which results in a change from aspartic acid to glycine; rs4789223, significantly associated with increased colon cancer risk in the haplotype analysis, is mapped at intron 8 and is not coding. There is moderate LD between these 2 SNPs; r-squared is 0.44.
Our study had 80% power to detect ORs ranging from approximately 1.34 to 1.40 given minor allele frequencies between 0.43 and 0.23, respectively, and thus this could explain the lack of definitive evidence of single SNP significance. Since haplotype analyses have better power to detect association when LD is taken into consideration, this may explain why the haplotype analyses were significant but not the SNPs alone [11]. If these results are replicated, the haplotype may harbor an untyped variant that could affect susceptibility.
In summary, we found that a haplotype in RECQL5 tagged by rs4789223 or other variants in LD with this SNP is associated with colon cancer in a case control study. Our results in combination with emerging evidence from model systems support RECQL5 as a novel susceptibility for colon cancer, and are worthy of further replication.
Grant Support
This research was supported by a Damon Runyon Cancer Research Foundation Clinical Investigator Award (CI-8) (LL), the Case Center for Transdisciplinary Research on Energetics and Cancer (U54 CA-116867-01) (LL), National Cancer Institute K22 Award (K22 CA120545-01) (LL), National Cancer Institute R25 Award (R25 CA094186) (CLT), State of Ohio Biomedical Research and Technology Transfer Commission (GC).
Financial Disclosures
None
| References | ▴Top |
- Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9(9):644-654.
pubmed doi - Hu Y, Lu X, Luo G. Effect of Recql5 deficiency on the intestinal tumor susceptibility of Apc(min) mice. World J Gastroenterol. 2010;16(12):1482-1486.
pubmed doi - Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM,
et al . RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21(23):3073-3084.
pubmed doi - Li L, Plummer SJ, Thompson CL, Tucker TC, Casey G. Association between phosphatidylinositol 3-kinase regulatory subunit p85alpha Met326Ile genetic polymorphism and colon cancer risk. Clin Cancer Res. 2008;14(3):633-637.
pubmed doi - Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25(2):115-121.
pubmed doi - Dudbridge F. 2006. UNPHASED user guide. In M. B. U. Technical Report 2006/5, editor, Cambridge, UK.
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I,
et al . Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426-1435.
pubmed doi - Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54(3):443-452.
pubmed doi - Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27(18):3762-3769.
pubmed doi - Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000;28(7):1647-1655.
pubmed doi - Fallin D, Cohen A, Essioux L, Chumakov I, Blumenfeld M, Cohen D, Schork NJ. Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer's disease. Genome Res. 2001;11(1):143-151.
pubmed doi
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.


