| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website http://www.gastrores.org |
Original Article
Volume 2, Number 6, December 2009, pages 311-316
No Association of Obesity and Type 2 Diabetes Mellitus Related Genetic Variants With Colon Cancer
Cheryl L. Thompsona, b, c, Sarah J. Plummerd, Thomas C. Tuckere, Graham Caseyd, Li Lia, b, c, f
aDepartments of Family Medicine, Case Western Reserve University/University Hospitals of Cleveland, 10900 Euclid Ave, Cleveland, OH 44106, USA
bEpidemiology and Biostatistics, Case Western Reserve University/University Hospitals of Cleveland, 10900 Euclid Ave, Cleveland, OH 44106, USA
cThe Case Center for Transdisciplinary Research on Energetics and Cancer, Case Comprehensive Cancer Center, 10900 Euclid Ave, Cleveland, OH 44106, USA
dDepartment of Preventive Medicine, University of Southern California, 1975 Zonal Ave, Los Angeles, CA 90089, USA
eCancer Control Program, University of Kentucky, 2365 Harrodsburg Rd, Lexington, KY 40504, USA
fCorresponding author: Case Western Reserve University, 10900 Euclid Ave, UCRC II 306, Cleveland, OH 44106, USA
Manuscript accepted for publication November 18, 2009
Short title: Association of Colon Cancer With Diabetes and Obesity
doi: https://doi.org/10.4021/gr2009.11.1323
| Abstract | ▴Top |
Background: Obesity and type 2 diabetes mellitus (T2D) are known risk factors for colon cancer. Recent reports from a number of genome wide association studies (GWAS) have identified several single nucleotide polymorphisms (SNPs) associated with obesity and T2D. Here we tested the hypothesis that these SNPs may also be associated with risk of colon cancer.
Methods: We genotyped nine SNPs reported in GWAS of obesity and/or T2D, including SNPs in HHEX, KCNJ11, SLC30A8, FTO, CDKN2, CDKAL1, TCF2, and the rs9300039 SNP in an intergenic region, in 561 colon cancer cases and 721 population controls.
Results: None of these SNPs were statistically significantly associated with colon cancer in our sample.
Conclusions: Overall, these results suggest that these obesity and T2D genetic susceptibility loci are unlikely to influence the risk of colon cancer.
Keywords: Obesity; Type 2 diabetes; Colon cancer; Single nucleotide polymorphisms; Association
| Introduction | ▴Top |
Obesity is now considered as a ‘probable’ cause for colon cancer [1], and insulin resistance with resulting metabolic perturbation is believed to be the key mechanism underlying the obesity-colon cancer link [2, 3]. Colon cancer and type 2 diabetes mellitus (T2D) share a remarkably similar risk factor profile, including obesity, high energy intake, a Western diet and physical inactivity. In addition, T2D itself is directly associated with colon cancer [2]. It is thus plausible that genetic susceptibility loci for obesity and T2D may also predispose to the development of colon cancer.
Genome-wide association studies (GWAS) have recently identified a number of novel single nucleotide polymorphisms (SNPs) that confer susceptibility to T2D [3-7] or obesity [8-11]. Although the functionality of these genetic variants is largely unknown, many of the genes in which the SNPs are located are known to be oncogenic or involved in molecular pathways implicated in carcinogenesis. For example, HHEX encodes a transcription factor in the homeobox family, and there is evidence that this gene can act as an oncogene [12]. CDKN2 is a well known tumor suppressor gene [13] which stabilizes the tumor suppressor p53. Mutations of the CDKN2 gene have been reported in numerous tumor types [14]. TCF2 (HNF1B), like HHEX, encodes a homeobox transcription factor. TCF2 has been recognized as a key player in carcinogenesis, especially in prostate cancer [15, 16].
To date, whether these genetic variants may also be associated with risk of cancer has not been investigated. We therefore tested this hypothesis in a population-based case-control study of colon cancer.
| Materials and Methods | ▴Top |
Study population
The study population is described elsewhere [17]. In brief, we recruited 561 incident colon cancer cases and 721 population controls. Colon cancer cases were identified via the Kentucky Surveillance, Epidemiology and End Results (SEER) Cancer Registry within 6 months of diagnosis. Controls were recruited via random digit dialing. All individuals were asked to donate a sample of blood for genomic DNA. We used the National Cancer Institute Colon Risk Factor Questionnaire (http://epi.grants.cancer.gov/documents/CFR/center_questionnaires/Colon/LA/ColonRiskFactor_USC.pdf) to collect complete lifestyle and behavioral risk factors, including demographic variables, family history of colorectal cancer and non-steroidal anti-inflammatory drug (NSAID) use. Body mass index was calculated based on self-reported height and weight. The response rate was approximately 72% cases, and 63% for eligible controls. The study was approved by the institutional review boards of Case Western Reserve University/University Hospitals Case Medical Center, the University of Kentucky, Lexington, and the University of Southern California, Los Angeles.
Selection of SNPs and genotyping
We selected nine SNPs previously identified to be associated with obesity (rs9939609 and rs8050136 in FTO) [8, 11] or T2D (rs1111875 in HHEX, rs5219 in KCNJ11, rs9300039, rs13266634 in SLC30A8, rs10811661 in CDKN2, rs7754840 in CDKAL1 and rs4430796 in TCF2) [3-7], as reported in GWAS through September 2007. All SNPs were genotyped by the TAQMAN assay using predesigned probe/primer sets from Applied Biosystems. Two percent of samples were repeated giving 100% concordance.
Statistical analyses
We first tested the fit of each SNP to Hardy-Weinberg proportions using a Chi-square goodness of fit. Since cases were specifically ascertained to a disease we hypothesize would be associated with these SNPs, we only used controls in this analysis.
To examine the association of each SNP with colon cancer, we initially assessed the univariate (individual SNP) association via a chi-square test. We further obtained odds ratios (OR) via a logistic regression accounting for age, sex and race (base model) as well as after fully adjusting for age, sex, race, BMI, non-steroidal anti-inflammatory drug (NSAID) use, and family history of colorectal cancer (full model). In the logistic regressions, we tested three genetic models (dominant, recessive or additive). For the additive model, each individual was coded as 0, 1 or 2, representing the number of risk (less frequent) alleles they possessed for each SNP. Dominant models were coded as 1 if the individual possessed 1 or 2 copies of the risk allele, 0 otherwise, whereas the recessive models were coded as 1 only if the individual possessed two copies of the risk allele, and 0 otherwise. To address possible spurious associations resulting from population stratification, we also performed analyses restricted to Caucasians only. SAS Version 9.1 (SAS Institute, Inc., Cary, North Carolina) was used to calculate all statistics. An α = 0.05 was used to declare statistical significance.
| Results | ▴Top |
On average, cases were older and more likely to be male than controls (Table 1). Compared to controls, cases had a higher mean BMI (p = 0.0077), were more likely to report a family history of colorectal cancer (24.0% vs. 15.4%, p = 0.0014), more likely to be diagnosed with diabetes (20.9% vs. 15.6%, p = 0.019), and less likely to regularly use NSAIDs (p = 0.16).
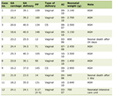 Click to view | Table 1. Descriptive Characteristics of Kentucky Colon Cancer Study Population |
All SNPs conformed to Hardy-Weinberg proportions among the controls (p > 0.05). We found no statistically significant association for any of the SNPs in univariate analyses (Table 1). After adjustment for potential confounders, all associations remained non-significant (Table 2). When the analyses were restricted to Caucasians, we found very similar results. For brevity, we have only presented results from the entire sample here.
 Click to view | Table 2. Descriptive Characteristics of Kentucky Colon Cancer Study Population |
| Discussion | ▴Top |
In this study, we found no association with colon cancer risk of a number of SNPs identified through T2D or obesity GWAS. Although many of the SNPs discovered through GWAS have been noted to have small effect sizes, we have at least 80% power to detect an odds ratio as low as 1.26, which is typical of most variants identified through GWAS, using the average minor allele frequency observed in these SNPs (33%) and assuming an additive model. Most of the SNPs included here are common variants with a relatively high frequency (> 10%) of the minor allele homozygotes. Thus it is unlikely that lack of statistical power could have obscured true association for these genetic variants. However, caution needs to be exercised for the KCNJ11 rs930039 and CDNK2 rs10811661 SNPs, for which the frequencies of the minor allele homozygotes are fairly low, limiting our power to detect an effect of these SNPs that truly follows a recessive mode of inheritance. Furthermore, we were able to replicate the association of the CDKAL1 rs7754840 SNP with self-reported T2D in this population, which compared to the most common GG genotype, individuals in our population with either the CG or CC genotype showed evidence of an increased risk of T2D (ORCG = 1.53, 95% CI = 1.08-2.16; ORCC = 1.63, 95% CI = 0.97-2.73; pdom = 0.0095 and padd = 0.015), lending credibility to our results.
There are limitations to our study. After we launched this project, other SNPs were identified in obesity or T2D GWAS, including MCR4 [10], TCF7L2 [3, 4] and EXT2 [7], but were not included in our study. In addition, population stratification can bias genetic association studies and produce spurious findings [18], and we only crudely accounted for potential population stratification by adjusting for self-reported race. However, given our null findings, population stratification does not appear to be an issue in our sample. Our study sample represents the general population in Kentucky, which is predominantly Caucasian. The small number of participants other than Caucasians (< 6%) limits our ability to estimate effects in other races.
Furthermore, we did not adjust for multiple testing in this study, but instead, examined these SNPs as novel candidate variants and tested a priori hypothesis of the existence of an association with colon cancer. Further adjustment for multiple testing would not alter our conclusion of no association with any of these SNPs and colon cancer.
Overall, we do not find evidence of shared genetic risk factors for colon cancer with obesity or T2D. However, the strong epidemiological and biological evidence for common underlying mechanisms influencing T2D and cancer warrants further investigation.
Grant Support
This research was supported by a Damon Runyon Cancer Research Foundation Clinical Investigator Award (CI-8), an R25 training grant from the National Cancer Institute (R25T CA094186), a K07 career development award from the National Cancer Institute (K07 CA136758-01A1), the Case Center for Transdisciplinary Research on Energetics and Cancer (1U54 CA-116867-01), and a National Cancer Institute K22 Award (1K22 CA120545-01).
Financial Disclosures
None of the authors have any financial conflicts with this manuscript.
| References | ▴Top |
- Panel WCRFW. Food, nutrition, physical activity, and the prevention of cancer: A global perspective. Washington, DC: WCRF/American Institute of Cancer Research; 2007.
- Giovannucci E. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am J Clin Nutr. 2007;86(3):s836-842.
pubmed - Meigs JB, Manning AK, Fox CS, Florez JC, Liu C, Cupples LA, Dupuis J. Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet. 2007;8 Suppl 1:S16.
pubmed - Salonen JT, Uimari P, Aalto JM, Pirskanen M, Kaikkonen J, Todorova B, Hypponen J,
et al . Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81(2):338-345.
pubmed doi - Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ,
et al . Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331-1336.
pubmed doi - Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR,
et al . A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341-1345.
pubmed doi - Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P,
et al . A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881-885.
pubmed doi - Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR,
et al . A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889-894.
pubmed doi - Liu YJ, Liu XG, Wang L, Dina C, Yan H, Liu JF, Levy S,
et al . Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet. 2008;17(12):1803-1813.
pubmed doi - Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M,
et al . Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(6):768-775.
pubmed doi - Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S,
et al . Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115.
pubmed doi - George A, Morse HC
3rd , Justice MJ. The homeobox gene Hex induces T-cell-derived lymphomas when overexpressed in hematopoietic precursor cells. Oncogene. 2003;22(43):6764-6773.
pubmed doi - Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264(1):42-55.
pubmed doi - Pollock PM, Pearson JV, Hayward NK. Compilation of somatic mutations of the CDKN2 gene in human cancers: non-random distribution of base substitutions. Genes Chromosomes Cancer. 1996;15(2):77-88.
pubmed doi - Frayling TM, Colhoun H, Florez JC. A genetic link between type 2 diabetes and prostate cancer. Diabetologia. 2008;51(10):1757-1760.
pubmed doi - Roose J, Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424(2-3):M23-37.
pubmed - Li L, Plummer SJ, Thompson CL, Tucker TC, Casey G. Association between phosphatidylinositol 3-kinase regulatory subunit p85alpha Met326Ile genetic polymorphism and colon cancer risk. Clin Cancer Res. 2008;14(3):633-637.
pubmed doi - Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361(9357):598-604.
pubmed doi
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.


